NOVINKA - PROTEOME DISCOVERER
ážení proteomici,
unikátny
softvér Proteome Discoverer (PD) v. 2.4
bol uvedený prednedávnom na trh a my Vám prinášame stručné zhrnutie, ako ho získať
či upgradovať Vašu súčasnú verziu. PD teraz využíva rovnaký licenčný mechanizmus
ako BioPharma Finder alebo Compound Discoverer. Ak si objednáte Proteome Discoverer 2.4,
dostanete email od ThermoMSLicensing@thermofisher.com s odkazom na stiahnutie
SW a licencie zo servera Flexera. Pre verziu 2.4 už nebude dostupná tzv.
„maintenance licence“, kedy išlo po dobu jedného roka zdarma upgradovať
na najnovšiu vydanú verziu. Pre staršie verzie je táto špeciálna utilita stále k dispozícii.
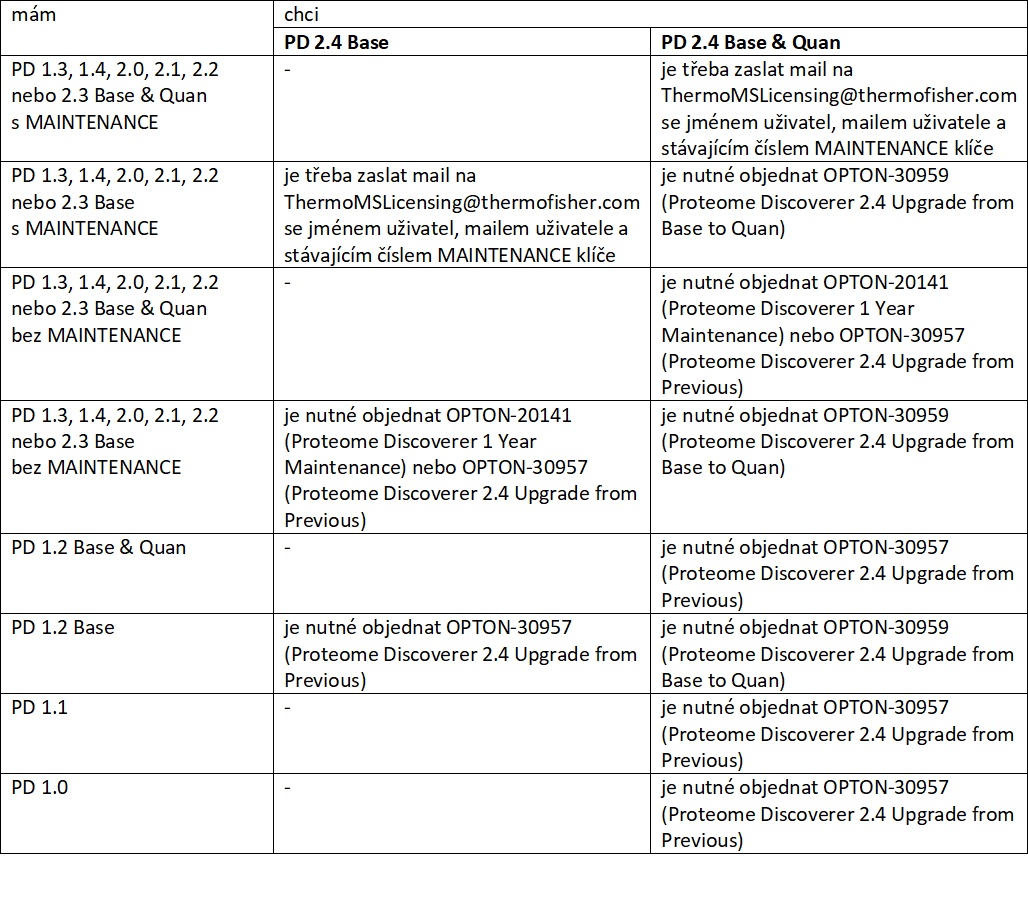
Úplne nový SW objednávajte pod kódom
OPTON-30945
(Proteome Discoverer 2.4 Base & Quan), upgrady pomocou nižšie uvedenej matice za použitia kódov
OPTON-30959
,
OPTON-20141
a OPTON-30957
. V prípade inej neštandardnej situácie či Vašej požiadavky, prosím, kontaktujte
našich obchodných zástupcov.
Proteome Discoverer 2.4 includes the
following new features
:
Precursor Detector node
Detects multiple precursor masses within isolation window
Searches the MS/MS spectrum multiple times using each precursor mass
Each precursor mass may produce a different PSM
For high resolution MS/MS data, use of the Precursor Detector node may increase protein ID’s up to 10% and unique peptides by 20%.
Scripting node
Enables users to call external scripts developed in R, Python, or other programming languages in a Proteome Discoverer workflow.
This will most commonly be used as a post-processing node to perform further data analysis after a workflow is complete.
Nodes can also be registered for use in Processing or Consensus workflows.
The scripting node currently can only access data from PD result tables (e.g. Proteins, Peptide Groups, etc.). Raw MS data and study information are inaccessible.
Please consult the scripting node section of the help for more information about the node. Examples of scripts in R, Python, and Java are included on the USB media.
Improved TMTpro 16plex support
Default TMTpro 16plex modification and quantification method
New table for TMTpro 16plex correction factors
New default workflow for TMTpro 16plex quantification
Note: Users of previous versions of PD can import the TMTpro 16plex quantification method and the TMTpro 16plex modification to analyze such data in that version. At the time of publication of this PDI, these files were not available to download but should be from either the Thermo Pierce website or the Flexera download site in the near future.
New Flexera licensing
The licensing mechanism has been changed to the same system as Compound Discoverer 3.1 and Biopharma Finder.
Updated license dialog
Users can now deactivate licenses to move their installation to a new PC.
Protein Annotation node
The Protein Annotation node is now tied to the Proteome Discoverer base license. Users will no longer need an active maintenance license to use this node.
Updated Familiarization Guide
Familiarization exercise 3 updated to include Precursor Detector node.
ProSightPD 3.0 (see ProSightPD 2.0 PDI)
XLinkX for PD2.4
FAIMS compensation voltage column in CSM tables
Can view target-decoy distributions for manual validation of the false discovery rate calculation.

